Predicting Hypoperfusion Lesion and Target Mismatch in Stroke from Diffusion-weighted MRI Using Deep Learning
Y. Yu, S. Christensen, J. Ouyang, F. Scalzo, D. Liebeskind, M. Lansberg, G. Albers, G. Zaharchuk
Radiology, 2022
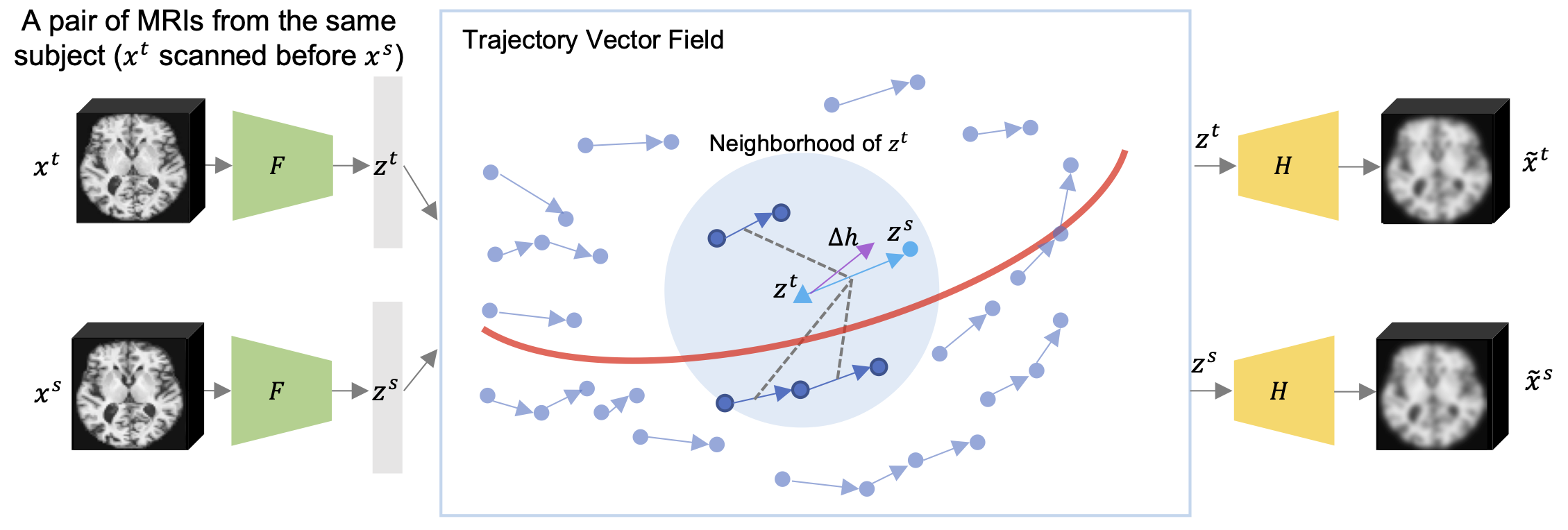
Self-supervised Learning of Neighborhood Embedding for Longitudinal MRI
J. Ouyang, Q. Zhao, E. Adeli, G. Zaharchuk, K. Pohl
Medical Image Analysis, 2022
Automated Detection of Arterial Landmarks and Vascular Occlusions in Patients with Acute Stroke Receiving Digital Subtraction Angiography Using Deep Learning
J. Khankari, Y. Yu, J. Ouyang, R. Hussein, H. Do, J. Heit, G. Zaharchuk
Journal of NeuroInterventional Surgery, 2022
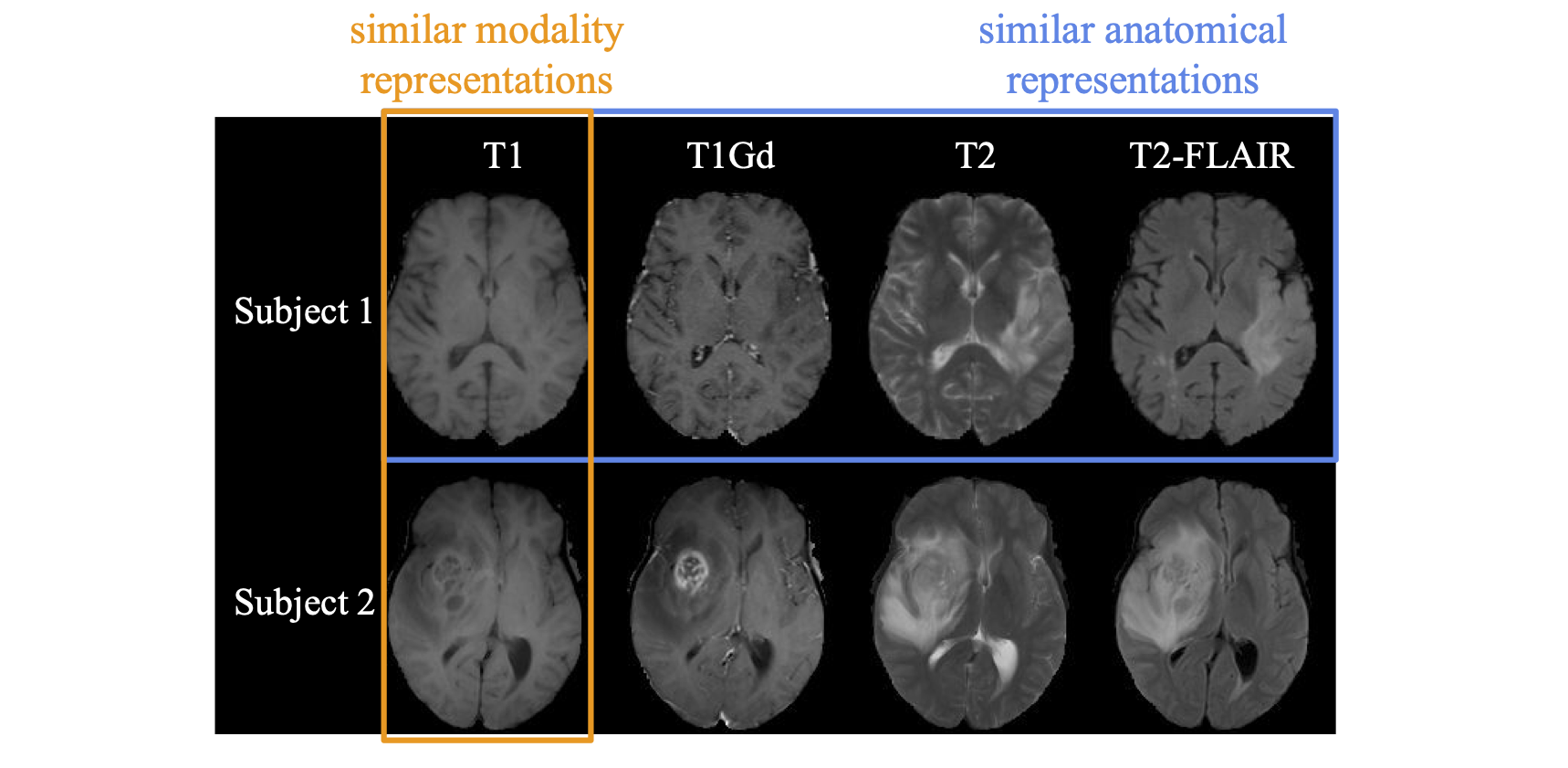
Disentangling Normal Aging from Severity of Disease via Weak Supervision on Longitudinal MRI
J. Ouyang, Q. Zhao, E. Adeli, G. Zaharchuk, K. Pohl
IEEE Transactions on Medical Imaging, 2022 [code]
Deep Learning Evaluation of Biomarkers from Echocardiogram Videos
J. Hughes, N. Yuan, B. He, J. Ouyang, ..., D. Ouyang, J. Zou
EBioMedicine, 2021
Tissue at Risk and Ischemic Core Estimation Using Deep Learning in Acute Stroke
Y. Yu, Y. Xie, T. Thamm, E. Gong, J. Ouyang, S. Christensen, M. Marks, M. Lansberg, G. Albers, G. Zaharchuk
American Journal of Neuroradiology, 2021 (Annual Lucien Levy Best Research Article Award)
Longitudinal Pooling & Consistency Regularization to Model Disease Progression from MRIs
J. Ouyang, Q. Zhao, E. Sullivan, A. Pfefferbaum, S. Tapert, E. Adeli, K. Pohl
Journal of Biomedical and Health Informatics (JBHI), 2020 [code]
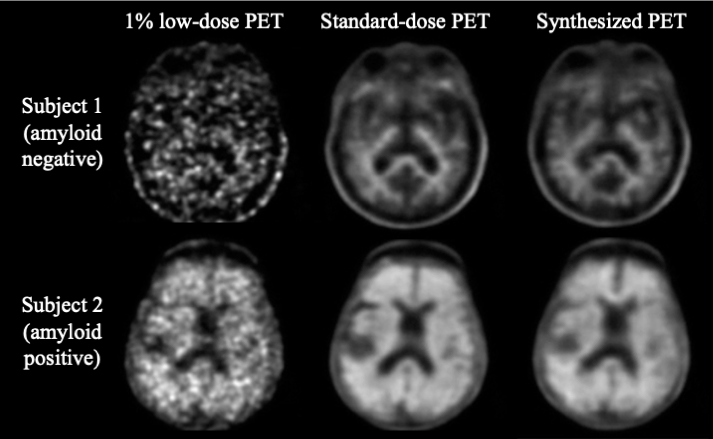
Generalization of Deep Learning Models for Ultra-low-count Amyloid PET/MRI Using Transfer Learning
K. Chen, M. Schurer, J. Ouyang, M. Koran, G. Davidzon, E. Mormino, S. Tieport, K. Hoffmann, O. Sabri, G. Zaharchuk, H. Barthel
European Journal of Nuclear Medicine and Molecular Imaging, 2020
Use of Deep Learning to Predict Final Ischemic Stroke Lesions From Initial Magnetic Resonance Imaging
Y. Yu, Y. Xie, T. Thamm, E. Gong, J. Ouyang, C. Huang, S. Christensen, M. Marks, M. Lansberg, G. Albers, G. Zaharchuk
Journal of the American Medical Association (JAMA) Network Open, 2020
Accurate Tissue Interface Segmentation via Adversarial Pre-segmentation of Anterior Segment OCT Images
J. Ouyang, T. Mathai, J. Galeotti
Biomedical Optics Express, 2019 [patent]
Ultra-low-dose Amyloid PET Reconstruction by Generative Adversarial Network with Feature Matching and Task-specific Perceptual Loss
J. Ouyang, E. Gong, K. Chen, J. Pauly, G. Zaharchuk
Medical Physics, 2019